🪢 Designing Protein Binder for Transmembrane Protein
- 1 min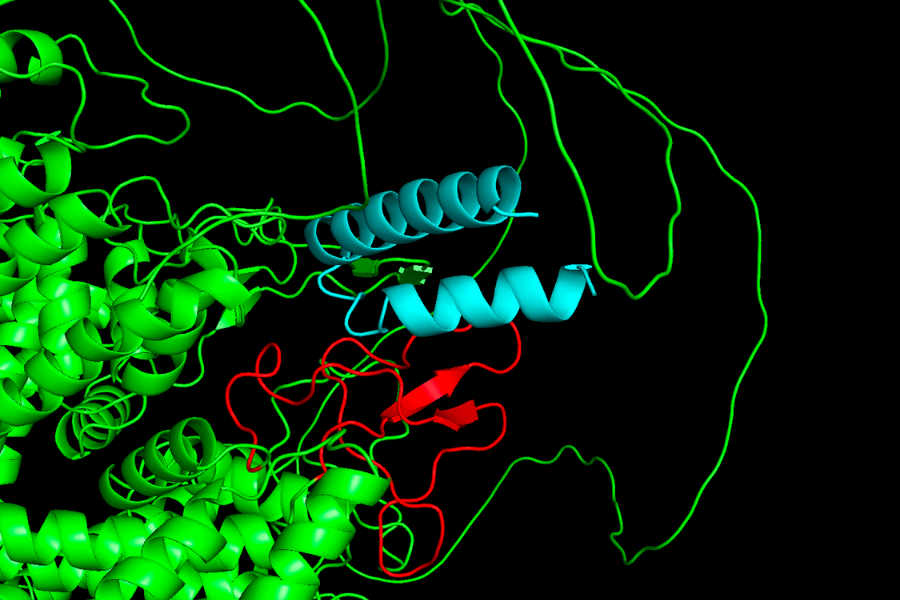
I had discovered a target protein using my former work at TenSixty Biosciences so far. This target protein was expressed only in the lungs of Small Cell Lung Cancer patients, and not in normal lungs. Now, I had to computationally develop a protein binder for this target protein.
The approach was simple- there already exists a Deep Learning tool called RFdiffusion, which takes in a protein identifier and the specific amino acids in it, corresponding to which we want to obtain a binder. It uses a guided diffusion model to find out new proteins, which can favorably bind to the target protein. The pae (protein alignment error) has to be minimised. If the pae score is around 10, one can go ahead with validating the obtained protein in lab. I was able to find a binder with pae score 11, which is shown in the figure.
If it works, then cancer drugs can be loaded on the binder protein designed, which will selectively bind to the target protein in the lungs of small cell lung cancer patients. The drug will thus be realeased only at the appropriate locations and specific drug targetting can be achieved.