Improving Polygenic Risk Prediction by Simulating Admixture
Dec 13, 2024
About:
This was me and my friend’s final project for the Statistical Genetics course at CMU during the Fall of 2024. I am proud of it because even though our analyses weren’t too many/complex, they were thorough, and we were able to present it effectively. I was mainly involved in simulating the admixed population part, and was happy to find that it displayed very similar patterns to actual admixed population.
Our motivation, goal and approach for the project were as follows:
- Motivation: current PRS methods only focus on individuals with distinct primary ancestry. Accounting for mosaic structure can increase power.
- Goal: Increase power of PRS methods by accounting for local ancestry information alongside individual genotype data.
- Approach: Using HapMap3 dataset, predict local ancestry probabilities of real and simulated admixed genotype data. Then, use local ancestry information and individual genotype data to perform polygenic risk prediction for diseases with known correlation (or lack of) to admixture.
Results:
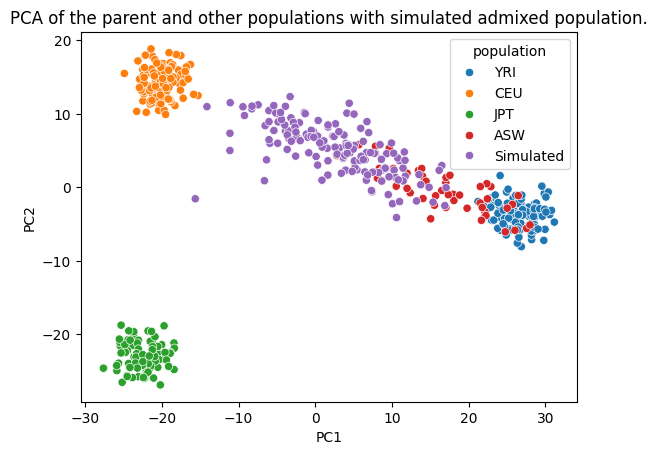
The PCA results clearly show that our simulated population is spread in the PCA plot between the composing populations just like the parent population. We used top 3k PCs to save computation, but the results were very similar with increasing PCs at that point.
STRUCTURE is a tool which allows identify k-components in each individual in a population, followed by clustering them and visualizing the components in each individual. The below plot shows that the actual admixed and simulated admixed populations are similar according to STRUCTURE results. Each vertical bar in the plot corresponds to a single individual, and the colors in the bar represent the components in that person for the k-categories. We chose k=3 here because there were 3 distinct populations here (the simulated and actual admixed pops were combinations of 2 of these).

Below figure shows how upon increasing distance between SNPs, the LD (linkage disequibrium) was still high in the actual admixed population and the simulated admixed population. This is a common behavior in admixed populations because of mixing of DNA in a mosaic fashion.

Links:
- github link to project: https://github.com/shashkat/Improving-Polygenic-Risk-Prediction-by-Simulating-Admixture
