🦠 Visualizing transmembrane proteins
- 1 min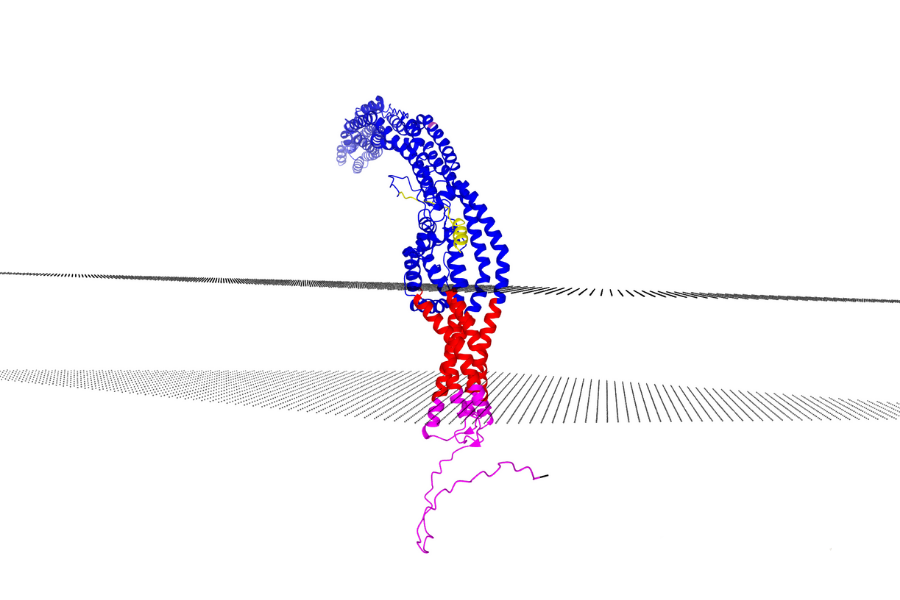
While working at TenSixty Biosciences, I was tasked with making a tool which could help visualize the intracellular and extracellular regions of transmembrane proteins. The reason this information was valuable because during drug designing, it is better that the target region of the target protein be extracellular. Thus, to ensure this and better visualize our target proteins, this tool was required.
I took assistance from an existing tool called DeepTMHMM (developed by some amazing people at Denmark Technical University), which uses Deep Learning on a sequence of amino acids and finds out the intracellular and extracellular regions in that. I took the information from DeepTMHMM and combined it with a template to view proteins in 3D. I colored the amino acids (blue-extracellular, yellow-signal, red-transmembrane and pink-intracellular) using the output from DeepTMHMM. To calculate the membrane location and show it, I used the locations of the amino acids where an intracellular/extracellular region was changing to transmembrane. The membrane is shown in the form of a bunch of water molecules on the appropriate plane.
Using this tool, we could find out the proteins of interest according to their extracellular regions and also the specific sites of interest in them for drug targetting. It proved to be very useful.